library(SIBER)
library(ggplot2)
library(ggExtra)
library(ggConvexHull)
library(HDInterval)
library(mcmcplots)
library(ggmcmc)
library(coda)
library(lattice)
library(MCMCvis)
library(bayesplot)3 Amplitudes and comparisons of the isotopic niches
3.1 Libraries
3.2 Data loading
data <- read.table("data/glm.csv", sep = ",", header = T)
colnames(data)[6:7] <- c("iso1", "iso2")3.3 “Global” SIBER:
3.3.1 Dataset arrangement
global <- subset(data, select = c(sp, iso1, iso2))
global$group <- factor(global$sp, labels = c(1:3))
global$community <- 1
global3.3.2 Basic plot
Parameters and basic information
global_colors <- c('#1f77b4', '#ff7f0e', '#2ca02c')
xlims <- c(min(global$iso2)-0.5, max(global$iso2)+0.5)
ylims <- c(min(global$iso1)-0.5, max(global$iso1)+0.5)
species <- unique(global$sp)Plot
glob_ellipses <- ggplot(data = global, aes(x = iso2, y = iso1, color = as.factor(group))) +
geom_point(alpha = 0.7) +
stat_ellipse(level = 0.4) +
geom_convexhull(fill = NA, linetype = "dashed", alpha = 0.05) +
scale_color_manual(values = global_colors, name = "Species", labels = species) +
scale_x_continuous(limits = xlims, breaks = c(seq(-18, -8, 2))) +
scale_y_continuous(limits = ylims, breaks = c(seq(10, 20, 2))) +
labs(title = element_blank(),
x = expression({delta}^13*C~'(\u2030)'),
y = expression({delta}^15*N~'(\u2030)')) +
theme_bw() +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.20, 0.17),
legend.background = element_blank())
# cairo_pdf("output/SIBER/global/global_ellipses.pdf",
# family = "Times", height = 3.5, width = 3.5)
glob_ellipses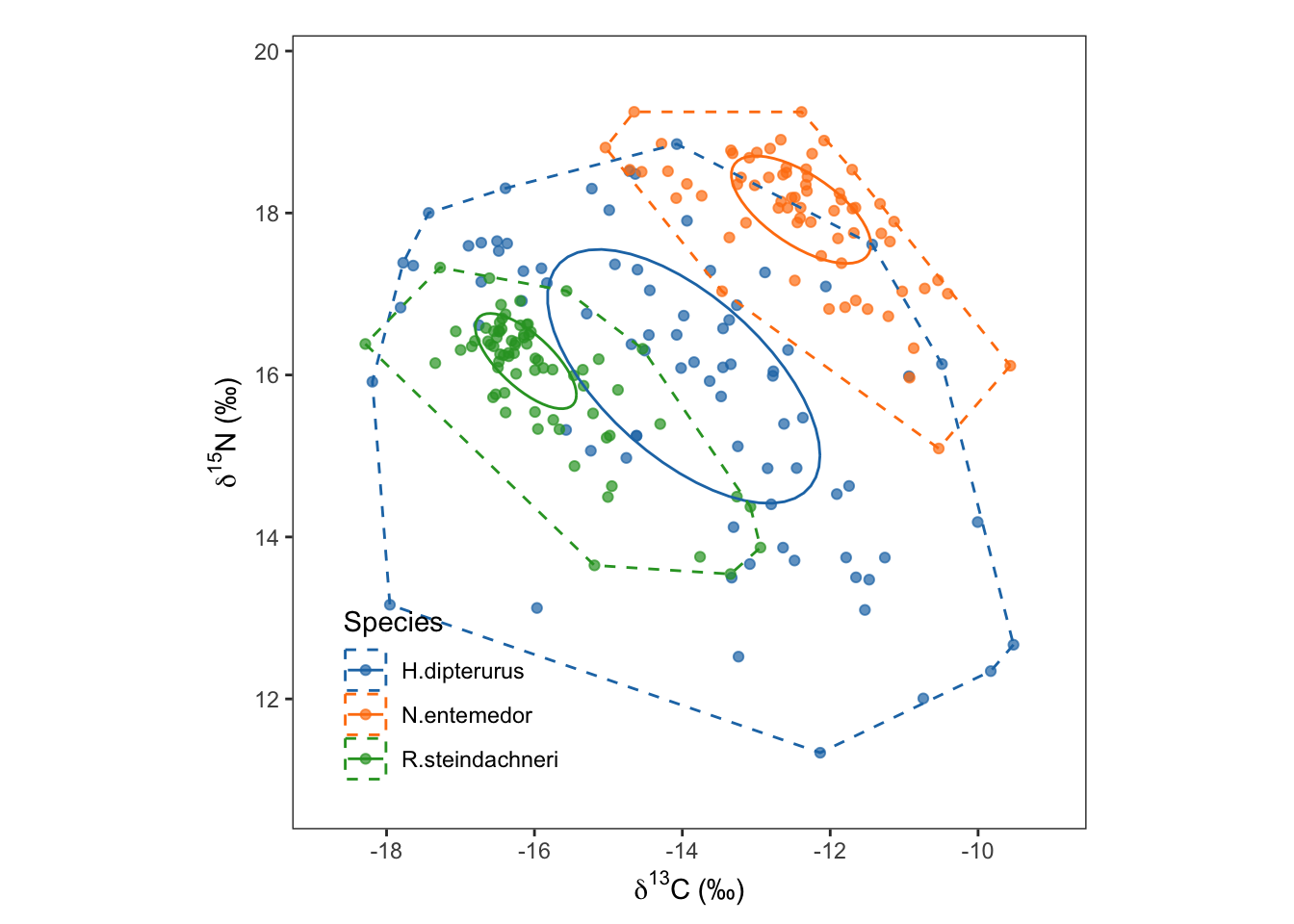
# dev.off()3.3.3 Fitting the Bayesian Bi-variate Normal models:
Creation of the SIBER object:
siber_glob <- createSiberObject(global[,2:5])Parameters of the MCMC:
parms <- list()
parms$n.iter <- 20000
parms$n.burnin <- 25000
parms$n.thin <- 1
parms$n.chains <- 3
parms$save.output <- TRUE
parms$save.dir <- paste(getwd(),"/output/SIBER/global", sep = "")Prior distributions:
priors <- list()
priors$R <- 1 * diag(2)
priors$k <- 2
priors$tau.mu <- 1.0E-3Sampling of the posterior distributions:
ellipses.posterior <- siberMVN(siber_glob, parms, priors)Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 81
Unobserved stochastic nodes: 3
Total graph size: 96
Initializing model
Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 69
Unobserved stochastic nodes: 3
Total graph size: 84
Initializing model
Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 74
Unobserved stochastic nodes: 3
Total graph size: 89
Initializing model3.3.3.1 Diagnostics:
Cycling through every model to extract information:
all_files <- dir(parms$save.dir, full.names = T)
model_files <- all_files[grep("jags_output", all_files)]
model_summary <- list()
for (i in seq_along(model_files)) {
load(model_files[i])
mcmc_list <- coda::as.mcmc(do.call("cbind", output))
model_summary[[i]] <- MCMCsummary(output,
params = "all",
probs = c(0.025, 0.975),
round = 3)
MCMCtrace(output,
params = "all",
iter = parms$n.iter,
Rhat = T,
n.eff = T,
ind = T,
type = "density",
open_pdf = F,
filename = sprintf("output/SIBER/global/MCMCtrace_%d", i))
}Autocorrelation plots:
# cairo_pdf("output/SIBER/global_autocorr.pdf",
# family = "Times", height = 10, width = 12)
gridExtra::grid.arrange(mcmc_acf(ellipses.posterior[1]) + labs(title = species[1]),
mcmc_acf(ellipses.posterior[2]) + labs(title = species[2]),
mcmc_acf(ellipses.posterior[3]) + labs(title = species[3]),
nrow = 3)
#dev.off()Summaries. Gelman-Rubick Statistics (Rhat) exactly equal to 1 and effective sample sizes > 2000 for every parameter:
for (i in seq_along(model_files)) {
load(model_files[i])
print(species[i])
print(model_summary[[i]])
}[1] "H.dipterurus"
mean sd 2.5% 97.5% Rhat n.eff
Sigma2[1,1] 1.025 0.165 0.751 1.393 1 60000
Sigma2[2,1] -0.557 0.132 -0.851 -0.331 1 59779
Sigma2[1,2] -0.557 0.132 -0.851 -0.331 1 59779
Sigma2[2,2] 1.025 0.165 0.753 1.392 1 58288
mu[1] 0.000 0.112 -0.221 0.220 1 31856
mu[2] 0.000 0.112 -0.220 0.221 1 31680
[1] "N.entemedor"
mean sd 2.5% 97.5% Rhat n.eff
Sigma2[1,1] 1.030 0.180 0.734 1.434 1 58420
Sigma2[2,1] -0.684 0.152 -1.025 -0.432 1 58657
Sigma2[1,2] -0.684 0.152 -1.025 -0.432 1 58657
Sigma2[2,2] 1.030 0.180 0.736 1.440 1 58409
mu[1] 0.001 0.121 -0.240 0.239 1 23172
mu[2] -0.001 0.122 -0.240 0.238 1 23062
[1] "R.steindachneri"
mean sd 2.5% 97.5% Rhat n.eff
Sigma2[1,1] 1.028 0.174 0.743 1.420 1 59100
Sigma2[2,1] -0.773 0.154 -1.119 -0.519 1 58438
Sigma2[1,2] -0.773 0.154 -1.119 -0.519 1 58438
Sigma2[2,2] 1.029 0.174 0.742 1.426 1 57750
mu[1] -0.002 0.118 -0.234 0.229 1 16294
mu[2] 0.002 0.118 -0.228 0.234 1 165153.3.3.2 Inferences
Generation of the ellipses:
sea_b <- siberEllipses(ellipses.posterior)
summary(sea_b) V1 V2 V3
Min. : 6.193 Min. :1.323 Min. :1.077
1st Qu.: 8.875 1st Qu.:1.971 1st Qu.:1.541
Median : 9.555 Median :2.136 Median :1.666
Mean : 9.636 Mean :2.156 Mean :1.681
3rd Qu.:10.311 3rd Qu.:2.318 3rd Qu.:1.804
Max. :15.689 Max. :3.721 Max. :3.167 Graphical comparisons. First, transform the object to a data.frame and melt it to long format:
sea_bt <- reshape2::melt(as.data.frame(sea_b))No id variables; using all as measure variablescolnames(sea_bt) <- c("sp", "SEAb")
head(sea_bt)Get the HDI for each species:
hdi <- as.data.frame(t(apply(sea_b, 2, FUN = HDInterval::hdi)))
hdi <- cbind(hdi, means = t(as.data.frame(t(apply(sea_b, 2, FUN = mean)))))
hdi["sp"] <- row.names(hdi)
hdiViolin plot with the posterior distributions of the SEAb:
seab_global <- ggplot() +
geom_violin(data = sea_bt, aes(x = sp, y = SEAb, fill = sp, color = sp),
show.legend = F, alpha = 0.3) +
geom_boxplot(data = sea_bt,
aes(x = sp, y = SEAb, fill = sp, color = sp),
alpha = 0.5, width = 0.05, notch = T, show.legend = F,
outlier.shape = NA, coef = 0) +
scale_fill_manual(values = global_colors) +
scale_color_manual(values = global_colors) +
geom_linerange(data = hdi,
aes(x = sp, ymin = lower, ymax = upper, color = sp),
show.legend = F) +
theme_bw()+
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.2, 0.15),
legend.background = element_blank()) +
labs(x = "Species",
y = expression("SEAb " ('\u2030' ^2) )) +
scale_x_discrete(labels = species)
# cairo_pdf("output/SIBER/global/global_seab.pdf",
# family = "Times", height = 3.5, width = 3.5)
seab_global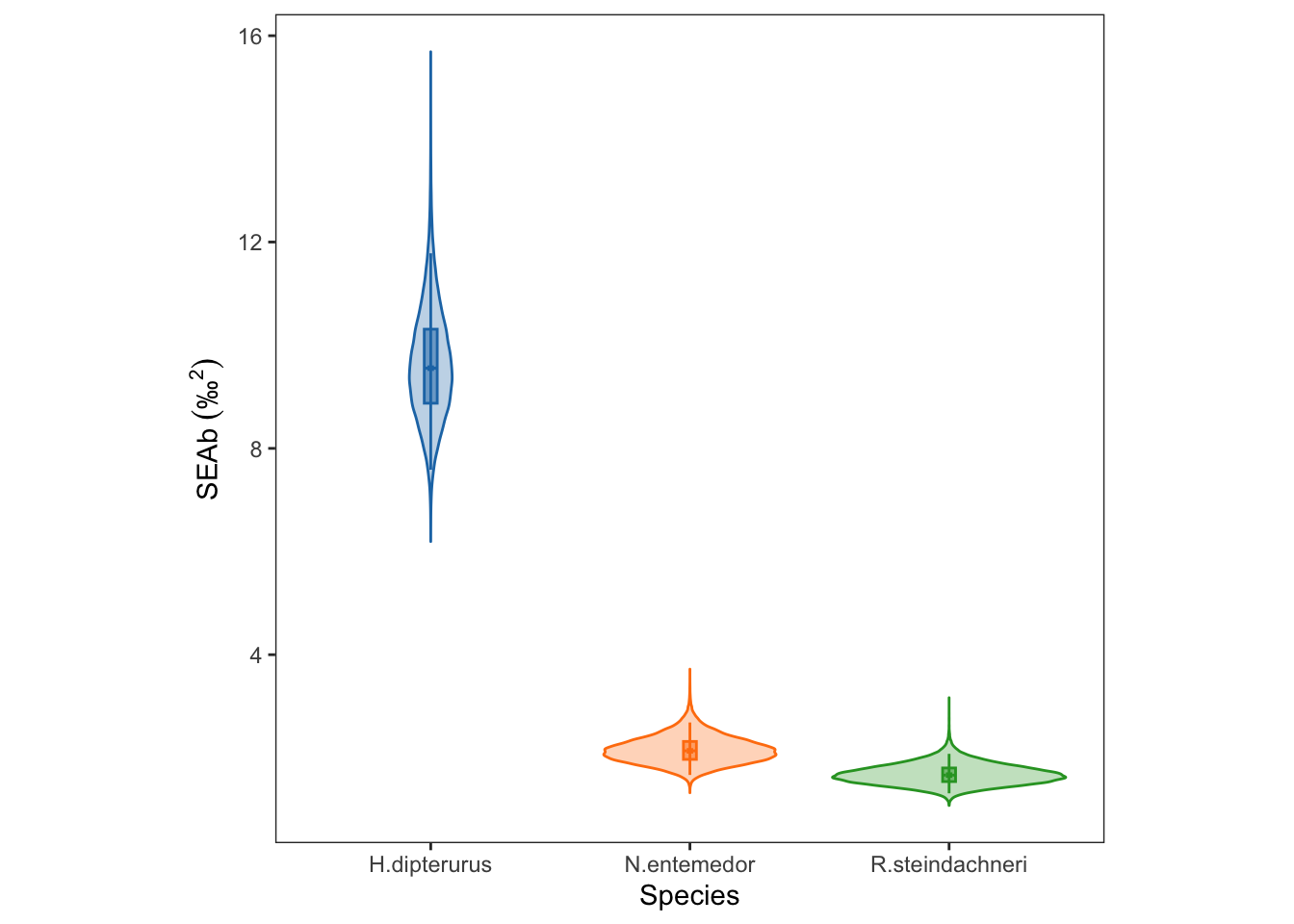
#dev.off()Posterior Differences:
# Define the comparisons
comps <- expand.grid(a = unique(as.integer(global$group)),
b = unique(as.integer(global$group)))
comps <- comps[(comps$a != comps$b & comps$b > comps$a), ]
comps <- comps[order(comps$a),]
# Put the results in different objects:
diffs <- data.frame()
diff_glob <- data.frame()
p_sup <- data.frame()
for (i in seq_along(comps$a)) {
a <- comps$a[i]
b <- comps$b[i]
comp <- paste(species[a], "-", species[b])
diff <- sea_b[, a] - sea_b[, b]
sup <- subset(diff, diff>0, select = diff)
diffs <- rbind(diffs,
data.frame(comp = comp,
diff = diff))
diff_glob <- rbind(diff_glob,
data.frame(comp = comp,
mean = mean(diff),
IQr = t(HDInterval::hdi(diff, credMass = 0.5)),
hdi = t(HDInterval::hdi(diff, credMass = 0.95))
)
)
p_sup <- rbind(p_sup,
data.frame(comp = comp,
p_sup = (length(sup)/length(diff))*100))
}
diff_glob$comp <- factor(diff_glob$comp, ordered = T)HDIs of differences
diff_glob["keys"] <- factor(c("HD-NE", "HD-RS", "NE-RS"), ordered = T)
diff_globProbabilities of differences:
p_supForestplot of the \(HDI_{95\%}\) of the differences
global_forest <- ggplot(data = diff_glob) +
geom_vline(xintercept = 0, linetype = "dashed", color = "#C9C8C8", size = 1.2) +
geom_linerange(aes(y = factor(comp, ordered = T),
xmin = IQr.lower, xmax = IQr.upper),
color = global_colors[1], size = 1) +
geom_linerange(aes(y = factor(comp, ordered = T),
xmin = hdi.lower, xmax = hdi.upper),
color = global_colors[1]) +
geom_point(aes(x = mean, y = factor(comp, ordered = T)),
color = global_colors[1],
fill = "white", size = 1.5, shape = 21) +
theme_bw() +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.2, 0.15),
legend.background = element_blank()) +
labs(title = expression("Differences in SEAB " ('\u2030' ^2)),
x = element_blank(),
y = element_blank())
# cairo_pdf("output/SIBER/global/global_forest.pdf",
# family = "Times", height = 4, width = 4)
global_forest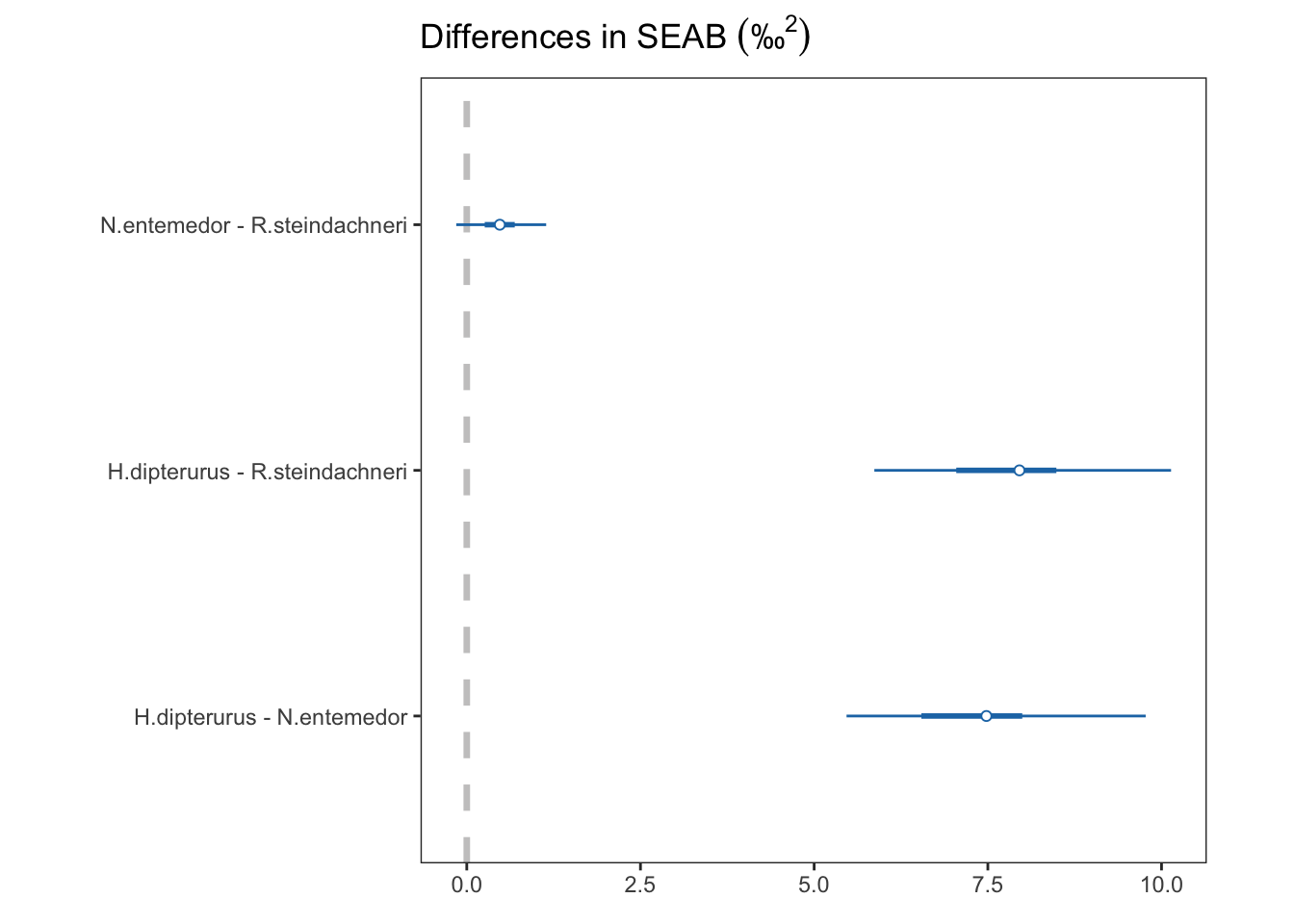
# dev.off()3.3.3.3 Eccentricity and theta
Eccentricities represent the elongation of the ellipse, that is, how far they are from a perfect circle (0 eccentricity). Lower values indicate a rounder ellipse, while higher values indicate a more elongated ellipse.
mat_ecc <- function(x) {
mat <- matrix(x, 2, 2)
ecc <- sigmaSEA(mat)$ecc
return(ecc)
}
mat_theta <- function(x){
mat <- matrix(x, 2, 2)
theta <- sigmaSEA(mat)$theta
return(theta)
}
ecc <- data.frame()
for (sp in seq_along(ellipses.posterior)) {
ecc <- rbind(ecc,
data.frame(sp = species[sp],
ecc = apply(ellipses.posterior[[sp]][,1:4], 1, mat_ecc),
theta = apply(ellipses.posterior[[sp]][,1:4], 1, mat_theta)))
}ec <- ecc %>% group_by(sp) %>% summarise(IQR.lower = hdi(ecc, credMass = 0.5)[1],
IQR.upper = hdi(ecc, credMass = 0.5)[2],
hdi.lower = hdi(ecc)[1],
hdi.upper = hdi(ecc)[2],
mean = mean(ecc))
ecggplot(data = ecc, aes(x = sp, y = ecc, group = sp, color = factor(sp))) +
geom_violin(fill = NA, show.legend = F) +
geom_point(aes(x = sp, y = mean), data = ec, show.legend = F) +
#geom_errorbar(aes(x = sp, ymin = IQ.lower, ymax = IQ.upper, group = sp), data = ag) +
geom_boxplot(width = 0.1, show.legend = F) +
theme_bw() +
labs(title = "SEAB Eccentricity",
x = element_blank(),
y = element_blank()) +
scale_color_manual(values = global_colors) +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.15, 0.1),
legend.background = element_blank())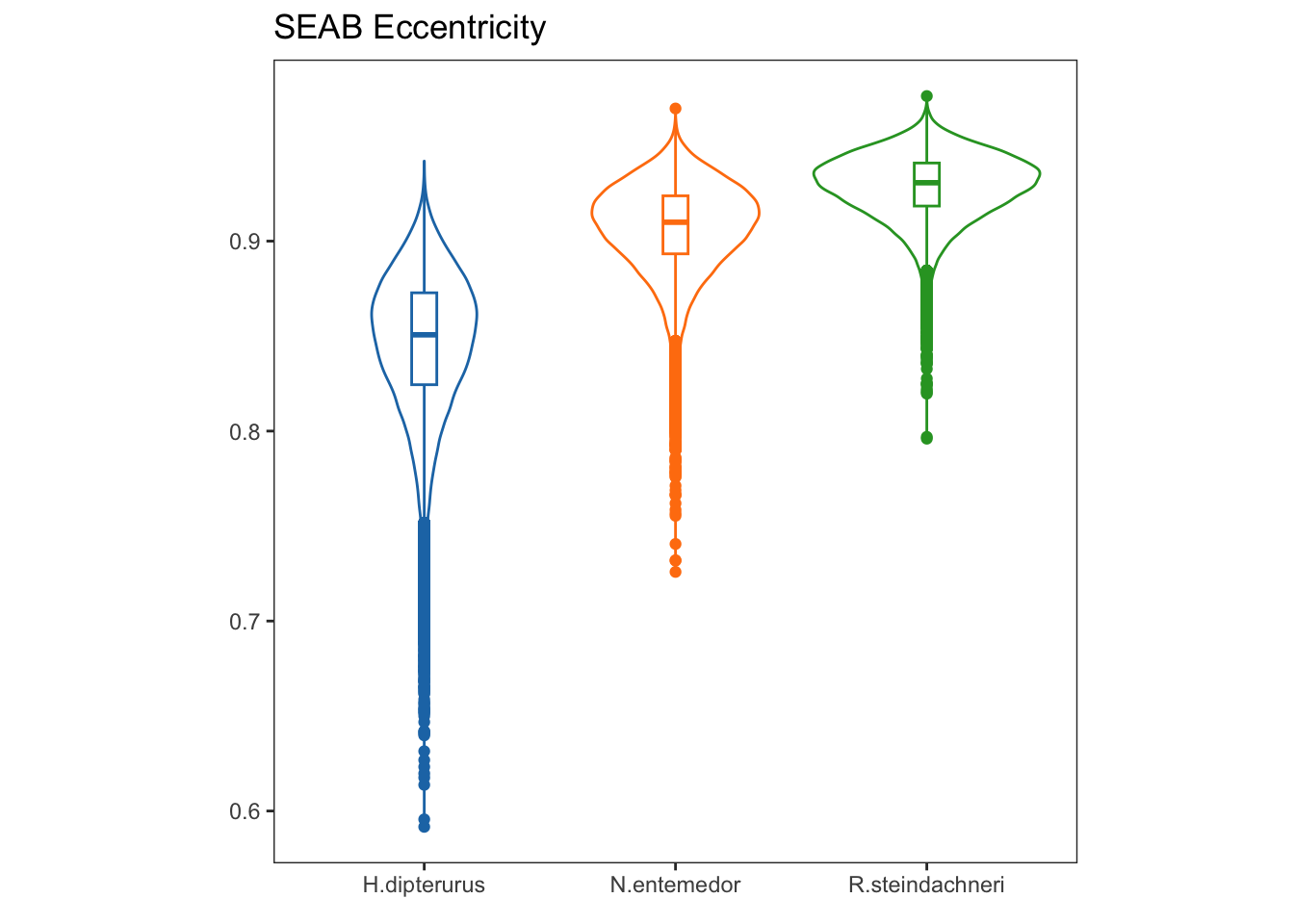
Theta, on the other hand, is the angle of the semi-major axis of the SEA and the x axis; i.e., the angle of the deformation. Values close to 0 show a higher dispersion in \(\delta^{13}C\), while values close to 90 show a higher dispersion in \(\delta^{15}N\):
th <- ecc %>% group_by(sp) %>% summarise(IQR.lower = hdi(theta, credMass = 0.5)[1],
IQR.upper = hdi(theta, credMass = 0.5)[2],
hdi.lower = hdi(theta)[1],
hdi.upper = hdi(theta)[2],
mean = mean(theta))
thggplot(data = ecc, aes(x = sp, y = theta, group = sp, color = factor(sp))) +
geom_violin(fill = NA, show.legend = F) +
geom_point(aes(x = sp, y = mean), data = ec, show.legend = F) +
#geom_errorbar(aes(x = sp, ymin = IQ.lower, ymax = IQ.upper, group = sp), data = ag) +
geom_boxplot(width = 0.1, show.legend = F) +
theme_bw() +
labs(title = "SEAB theta",
x = element_blank(),
y = element_blank()) +
scale_color_manual(values = global_colors) +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.15, 0.1),
legend.background = element_blank())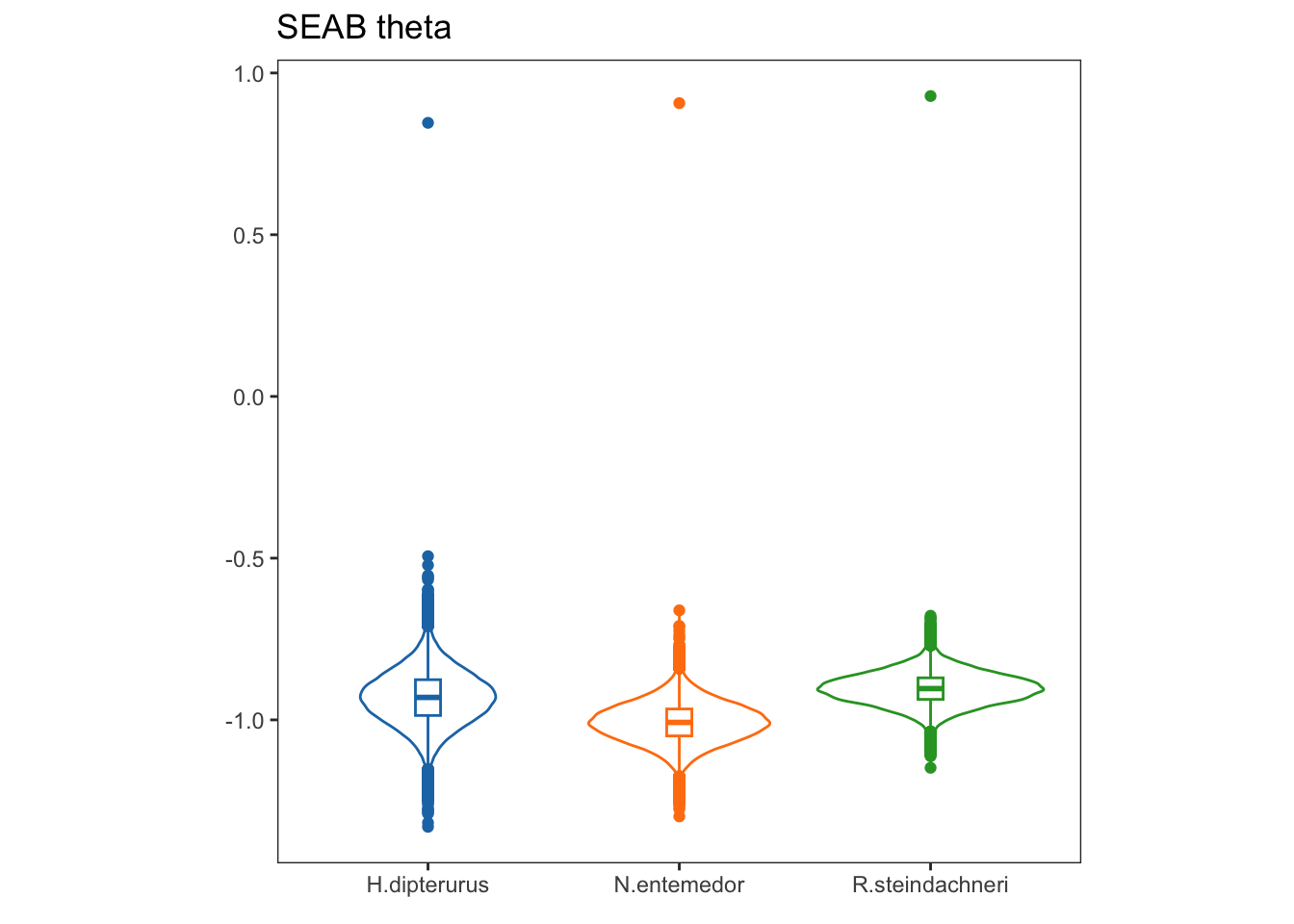
3.4 Intra-specific analyses
Basic dataset to extract every case:
full_set <- subset(data,
select = c(sp, temporada, sexo, estadio, iso1, iso2))
colnames(full_set) <- c("Species", "Season", "Sex", "Maturity", "iso1", "iso2")
head(full_set)3.4.1 Custom wrapper around SIBER:
custom_siber <- function(data, var_col, lbls, sp_col, colors, gp_label,
n.burnin = 30000, n.iter = 2000){
subdata <- data[,c(sp_col, var_col, "iso1", "iso2")]
subdata$group <- factor(subdata[,var_col], labels = seq_along(lbls))
subdata$community <- 1
species <- unique(data[, sp_col])
# Global directory for the variable
wd <- paste(getwd(), "/output/SIBER/", var_col, sep = "")
# If the directory does not exist, create it:
if (dir.exists(wd) == F) {dir.create(wd)}
# Empty lists to store the results:
siber_objs <- list()
ellipses <- list()
summaries <- list()
autocorr_plots <- list()
sea_b <- list()
sea_bt <- data.frame()
hdis <- data.frame()
ecc <- data.frame()
# Posterior differences
# Define the comparisons
comps <- expand.grid(a = unique(as.integer(subdata$group)),
b = unique(as.integer(subdata$group)))
comps <- comps[(comps$a != comps$b & comps$b > comps$a), ]
comps <- comps[order(comps$a),]
# Put the results in different objects:
diffs <- data.frame()
diff_glob <- data.frame()
p_sup <- data.frame()
# List with MCMC parameters:
parms <- list()
parms <- list()
parms$n.iter <- n.iter
parms$n.burnin <- n.burnin
parms$n.thin <- 1
parms$n.chains <- 3
parms$save.output <- TRUE
# List with prior parameters:
priors <- list()
priors$R <- 1 * diag(2)
priors$k <- 2
priors$tau.mu <- 1.0E-3
# Basic ellipses plot
ellipses <- ggplot(data = subdata, aes(x = iso2, y = iso1, color = as.factor(group))) +
geom_point(alpha = 0.5) +
stat_ellipse(level = 0.4) +
geom_convexhull(fill = NA, linetype = "dashed", alpha = 0.05) +
scale_color_manual(values = global_colors, name = gp_label, labels = lbls) +
scale_x_continuous(limits = xlims, breaks = c(-18, -14, -10)) +
scale_y_continuous(limits = ylims, breaks = c(12, 16, 20)) +
labs(title = element_blank(),
x = expression({delta}^13*C~'(\u2030)'),
y = expression({delta}^15*N~'(\u2030)')) +
theme_bw() +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
#legend.position = c(0.2, 0.15),
legend.background = element_blank()) +
facet_wrap(paste("~", sp_col, sep = ""))
cairo_pdf(paste(wd, "ellipses.pdf", sep = "/"), family = "Times", height = 2.7, width = 5.4)
print(ellipses)
dev.off()
# Loop through species:
for (i in seq_along(species)) {
# Output directory:
gpwd <- paste(wd, "/", species[i], sep = "")
if (dir.exists(gpwd) == F) {dir.create(gpwd)}
parms$save.dir <- gpwd
# SIBER objects
siber_objs[[species[i]]] <- createSiberObject(subdata[subdata[, sp_col] == species[i],3:6])
# Fitting the Bayesian models:
ellipses[[species[i]]] <- siberMVN(siber_objs[[i]], parms, priors)
# Diagnostics
## Get all the files in the directory
all_files <- dir(parms$save.dir, full.names = T)
model_files <- all_files[grep("jags_output", all_files)]
# Empty lists to store the results
summaries[[species[i]]] <- list()
autocorr_plots[[species[i]]] <- list()
# For each model:
for (j in seq_along(model_files)) {
# Load the model output
load(model_files[j])
# Create a list with the output
mcmc_list <- coda::as.mcmc(do.call("cbind", output))
# Store the summary for each model
summaries[[species[i]]][[j]]<- MCMCsummary(output,
params = "all",
probs = c(0.025, 0.975),
round = 3)
# Write the summary to a csv file
write.csv(summaries[[species[i]]][[j]],
paste(gpwd, sprintf("summary_%s.csv",
lbls[j]), sep = "/"),
row.names = T)
# Plot the trace in a pdf
MCMCtrace(output,
params = "all",
iter = parms$n.iter,
Rhat = T,
n.eff = T,
ind = T,
type = "density",
open_pdf = F,
#filename = paste(gpwd, sprintf("/MCMCtrace_%d", j), sep = ""))
filename = sprintf("MCMCtrace_%s", lbls[j]),
wd = gpwd)
# Store the autocorrelation plots
autocorr_plots[[species[i]]][[j]] <- mcmc_acf(ellipses[[species[i]]][j]) + labs(title = lbls[j])
} # End of models loop
# Autocorrelation plots for every model for every species
cairo_pdf(paste(gpwd, "/autocorr.pdf", sep = ""), family = "Times")
gridExtra::grid.arrange(grobs = autocorr_plots[[species[i]]], nrow = 2)
dev.off()
# Generation of the ellipses
sea_b[[species[i]]] <- siberEllipses(ellipses[[species[i]]])
# Fill the data.frame with the SEAb data
sea_bt <- rbind(sea_bt,
cbind(reshape2::melt(as.data.frame(sea_b[[species[i]]])),
sp_col = species[i]))
if (i == max(seq_along(species))) {colnames(sea_bt) <- c(var_col, "SEAb", sp_col)}
# Create a data.frame to store the hdis
# Calculate posterior means
means <- t(as.data.frame(t(apply(sea_b[[species[i]]], 2, FUN = mean))))
# Calculate hdis
hdi <- rbind(as.data.frame(t(apply(sea_b[[species[i]]], 2, FUN = HDInterval::hdi))))
# Put the results in a data.frame
temp <- data.frame(hdi, means, sp = species[i])
# Fill the data.frame with the results
hdis <- rbind(hdis, temp)
if (i == max(seq_along(species))) {hdis[var_col] <- paste("V", rep(seq_along(model_files)), sep = "")}
# Compute the posterior SEAb differences
for (k in seq_along(comps$a)) {
a <- comps$a[k]
b <- comps$b[k]
comp <- paste(lbls[a], "-", lbls[b])
diff <- sea_b[[species[i]]][, a] - sea_b[[species[i]]][, b]
sup <- subset(diff, diff>0, select = diff)
diffs <- rbind(diffs,
data.frame(comp = comp,
diff = diff,
sp = species[i]))
diff_glob <- rbind(diff_glob,
data.frame(comp = comp,
mean = mean(diff),
IQr = t(HDInterval::hdi(diff, credMass = 0.5)),
hdi = t(HDInterval::hdi(diff, credMass = 0.95)),
sp = species[i]
)
)
p_sup <- rbind(p_sup,
data.frame(comp = comp,
p_sup = (length(sup)/length(diff))*100,
sp = species[i]))
}
diff_glob$comp <- factor(diff_glob$comp, ordered = T)
}# End of species loop
# Violin plot of SEAb
seab <- ggplot() +
geom_violin(data = sea_bt, aes_string(x = var_col, y = "SEAb", fill = var_col, color = var_col),
show.legend = F, alpha = 0.3) +
geom_boxplot(data = sea_bt,
aes_string(x = var_col, y = "SEAb", fill = var_col, color = var_col),
alpha = 0.5, width = 0.05, notch = T, show.legend = F,
outlier.shape = NA, coef = 0) +
scale_fill_manual(values = global_colors) +
scale_color_manual(values = global_colors) +
# geom_linerange(data = hdi,
# aes_string(x = var_col, ymin = "lower", ymax = "upper", color = var_col),
# show.legend = F) + # Fix needed
theme_bw() +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.2, 0.15),
legend.background = element_blank()) +
labs(x = gp_label,
y = expression("SEAb " ('\u2030' ^2) )) +
scale_x_discrete(labels = lbls) +
facet_wrap(paste(".~", sp_col, sep = ""), ncol = 3, scales = "free_y")
cairo_pdf(paste(wd, "SEAb.pdf", sep = "/"), family = "Times", height = 4, width = 8)
print(seab)
dev.off()
print("All models have been run. Check your working directory for the results")
return(list(summaries = summaries,
sea_b = sea_b,
sea_bt = sea_bt,
hdis = hdis,
diffs = diffs,
diff_glob = diff_glob,
p_sup = p_sup,
ellips = ellipses))
}3.4.2 Sex
lbls <- c("Female", "Male")
sex_siber <- custom_siber(full_set, var_col = "Sex", lbls, sp_col = "Species", n.burnin = 7000,
colors = global_colors, gp_label = "Sex")Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 37
Unobserved stochastic nodes: 3
Total graph size: 52
Initializing model
Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 44
Unobserved stochastic nodes: 3
Total graph size: 59
Initializing modelNo id variables; using all as measure variablesCompiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 55
Unobserved stochastic nodes: 3
Total graph size: 70
Initializing model
Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 14
Unobserved stochastic nodes: 3
Total graph size: 29
Initializing modelNo id variables; using all as measure variablesCompiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 31
Unobserved stochastic nodes: 3
Total graph size: 46
Initializing model
Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 43
Unobserved stochastic nodes: 3
Total graph size: 58
Initializing modelNo id variables; using all as measure variablesWarning: `aes_string()` was deprecated in ggplot2 3.0.0.
ℹ Please use tidy evaluation ideoms with `aes()`[1] "All models have been run. Check your working directory for the results"Plots of ellipses:
sex_siber$ellips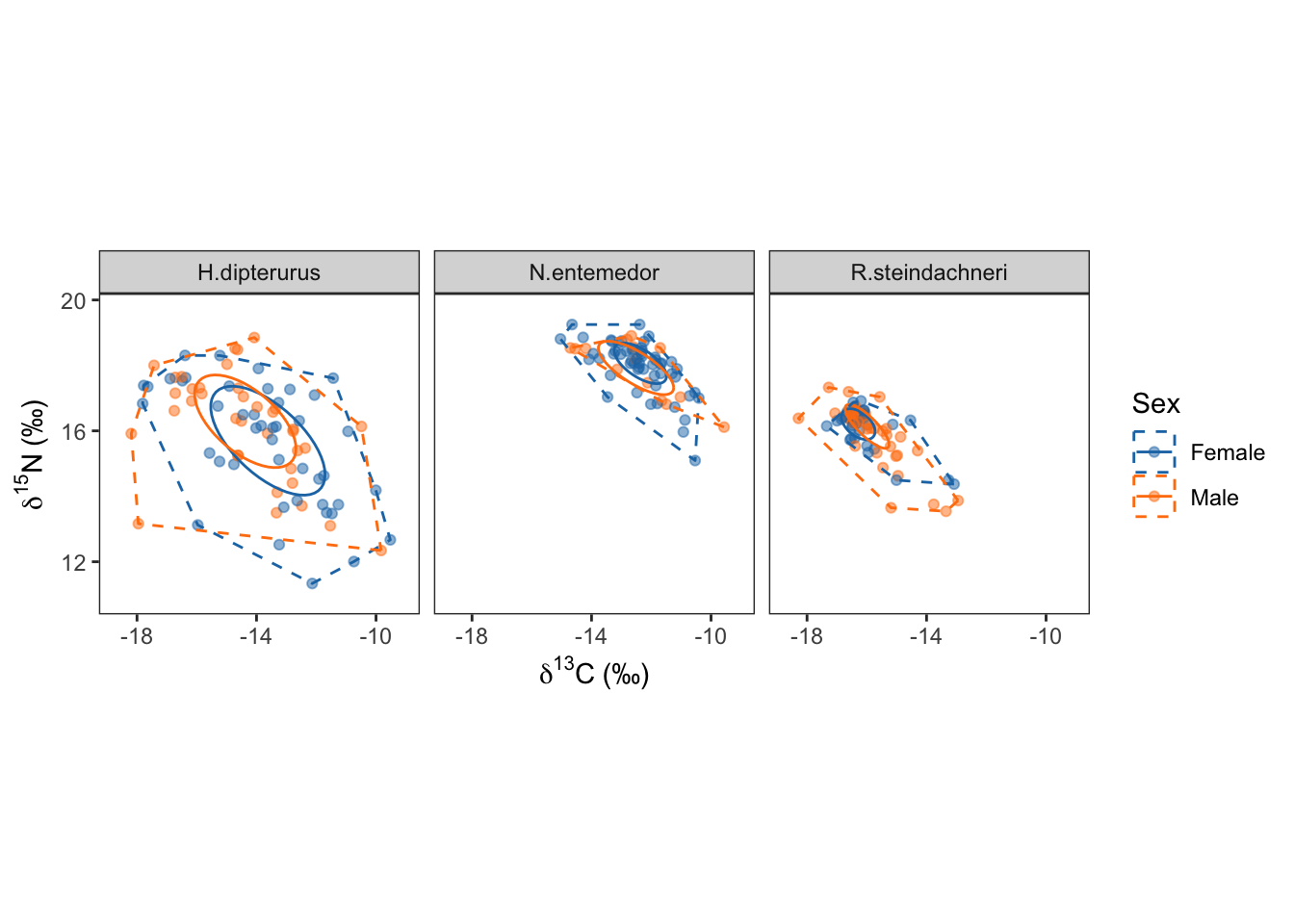
Eccentricities:
ecc <- data.frame()
for (sp in seq_along(species)) {
for (j in seq_along(lbls)) {
ecc <- rbind(ecc,
data.frame(sp = species[sp],
j = lbls[j],
ecc = apply(sex_siber$ellips[[species[sp]]][[j]][,1:4], 1, mat_ecc),
theta = apply(sex_siber$ellips[[species[sp]]][[j]][,1:4], 1, mat_theta)))
}
}ec <- ecc %>% group_by(sp, j) %>% summarise(IQR.lower = hdi(ecc, credMass = 0.5)[1],
IQR.upper = hdi(ecc, credMass = 0.5)[2],
hdi.lower = hdi(ecc)[1],
hdi.upper = hdi(ecc)[2],
mean = mean(ecc))`summarise()` has grouped output by 'sp'. You can override using the `.groups`
argument.ecggplot(data = ecc, aes(x = j, y = ecc, group = j, color = factor(j))) +
geom_violin(fill = NA, show.legend = F) +
#geom_point(aes(x = sp, y = mean), data = ag, show.legend = F) +
#geom_errorbar(aes(x = sp, ymin = IQ.lower, ymax = IQ.upper, group = sp), data = ag) +
geom_boxplot(width = 0.1, show.legend = F) +
theme_bw() +
labs(title = "SEAB Eccentricity",
x = element_blank(),
y = element_blank()) +
scale_color_manual(values = global_colors) +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.15, 0.1),
legend.background = element_blank()) +
facet_wrap(~sp)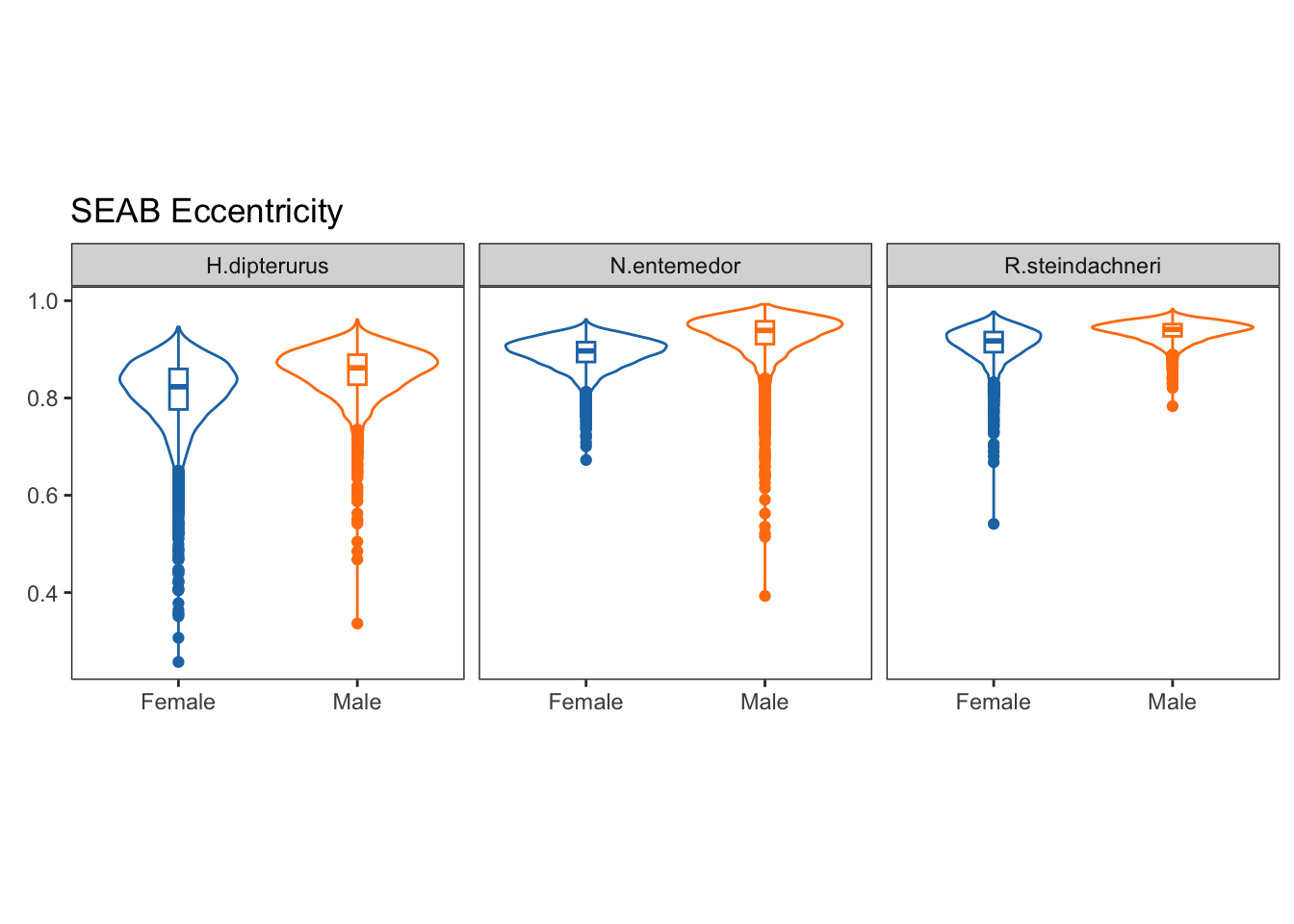
Theta:
th <- ecc %>% group_by(sp, j) %>% summarise(IQR.lower = hdi(theta, credMass = 0.5)[1],
IQR.upper = hdi(theta, credMass = 0.5)[2],
hdi.lower = hdi(theta)[1],
hdi.upper = hdi(theta)[2],
mean = mean(theta))`summarise()` has grouped output by 'sp'. You can override using the `.groups`
argument.thggplot(data = ecc, aes(x = j, y = theta, group = j, color = factor(j))) +
geom_violin(fill = NA, show.legend = F) +
#geom_point(aes(x = sp, y = mean), data = ag, show.legend = F) +
#geom_errorbar(aes(x = sp, ymin = IQ.lower, ymax = IQ.upper, group = sp), data = ag) +
geom_boxplot(width = 0.1, show.legend = F) +
theme_bw() +
labs(title = "SEAB theta",
x = element_blank(),
y = element_blank()) +
scale_color_manual(values = global_colors) +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.15, 0.1),
legend.background = element_blank()) +
facet_wrap(~sp)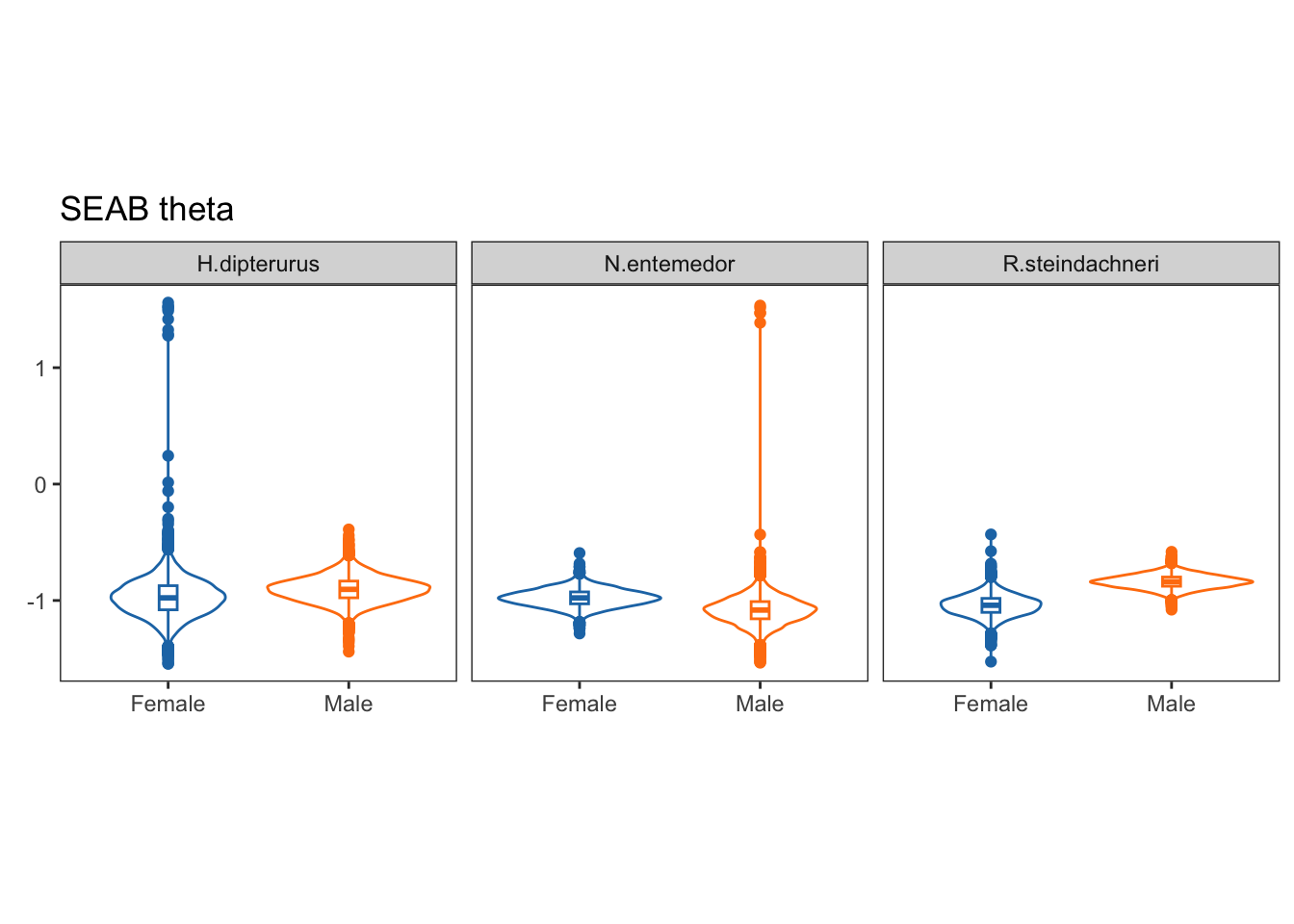
3.4.3 Maturity stages
lbls <- c("Adult", "Juvenile")
age_siber <- custom_siber(full_set, var_col = "Maturity", lbls, sp_col = "Species", n.burnin = 7000,
colors = global_colors, gp_label = "Age")Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 45
Unobserved stochastic nodes: 3
Total graph size: 60
Initializing model
Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 36
Unobserved stochastic nodes: 3
Total graph size: 51
Initializing modelNo id variables; using all as measure variablesCompiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 55
Unobserved stochastic nodes: 3
Total graph size: 70
Initializing model
Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 14
Unobserved stochastic nodes: 3
Total graph size: 29
Initializing modelNo id variables; using all as measure variablesCompiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 48
Unobserved stochastic nodes: 3
Total graph size: 63
Initializing model
Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 26
Unobserved stochastic nodes: 3
Total graph size: 41
Initializing modelNo id variables; using all as measure variables[1] "All models have been run. Check your working directory for the results"Plots of ellipses:
age_siber$ellips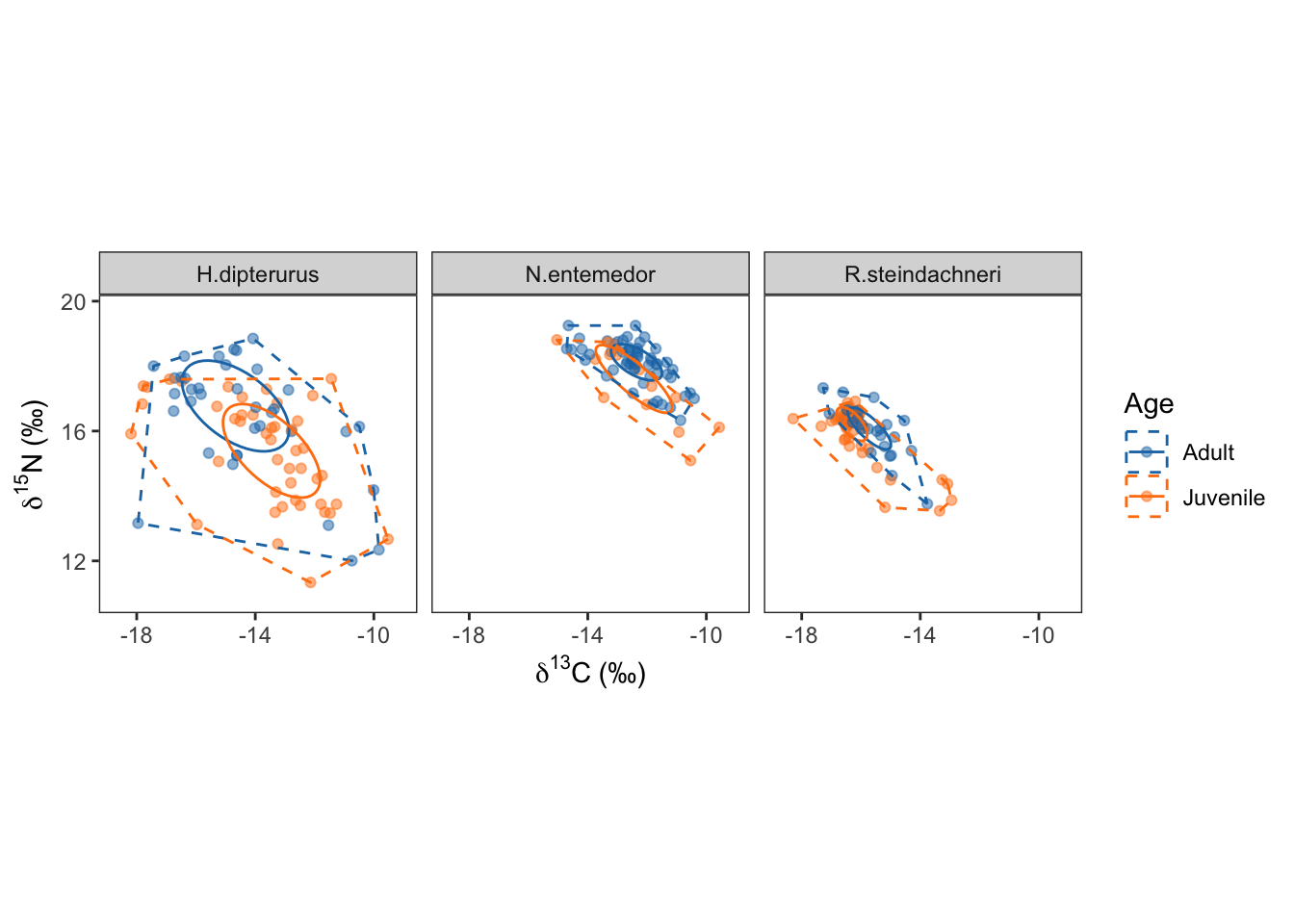
Eccenctricities:
ecc <- data.frame()
for (sp in seq_along(species)) {
for (j in seq_along(lbls)) {
ecc <- rbind(ecc,
data.frame(sp = species[sp],
j = lbls[j],
ecc = apply(age_siber$ellips[[species[sp]]][[j]][,1:4], 1, mat_ecc),
theta = apply(age_siber$ellips[[species[sp]]][[j]][,1:4], 1, mat_theta)))
}
}ec <- ecc %>% group_by(sp, j) %>% summarise(IQR.lower = hdi(ecc, credMass = 0.5)[1],
IQR.upper = hdi(ecc, credMass = 0.5)[2],
hdi.lower = hdi(ecc)[1],
hdi.upper = hdi(ecc)[2],
mean = mean(ecc))`summarise()` has grouped output by 'sp'. You can override using the `.groups`
argument.ecggplot(data = ecc, aes(x = j, y = ecc, group = j, color = factor(j))) +
geom_violin(fill = NA, show.legend = F) +
#geom_point(aes(x = sp, y = mean), data = ag, show.legend = F) +
#geom_errorbar(aes(x = sp, ymin = IQ.lower, ymax = IQ.upper, group = sp), data = ag) +
geom_boxplot(width = 0.1, show.legend = F) +
theme_bw() +
labs(title = "SEAB Eccentricity",
x = element_blank(),
y = element_blank()) +
scale_color_manual(values = global_colors) +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.15, 0.1),
legend.background = element_blank()) +
facet_wrap(~sp)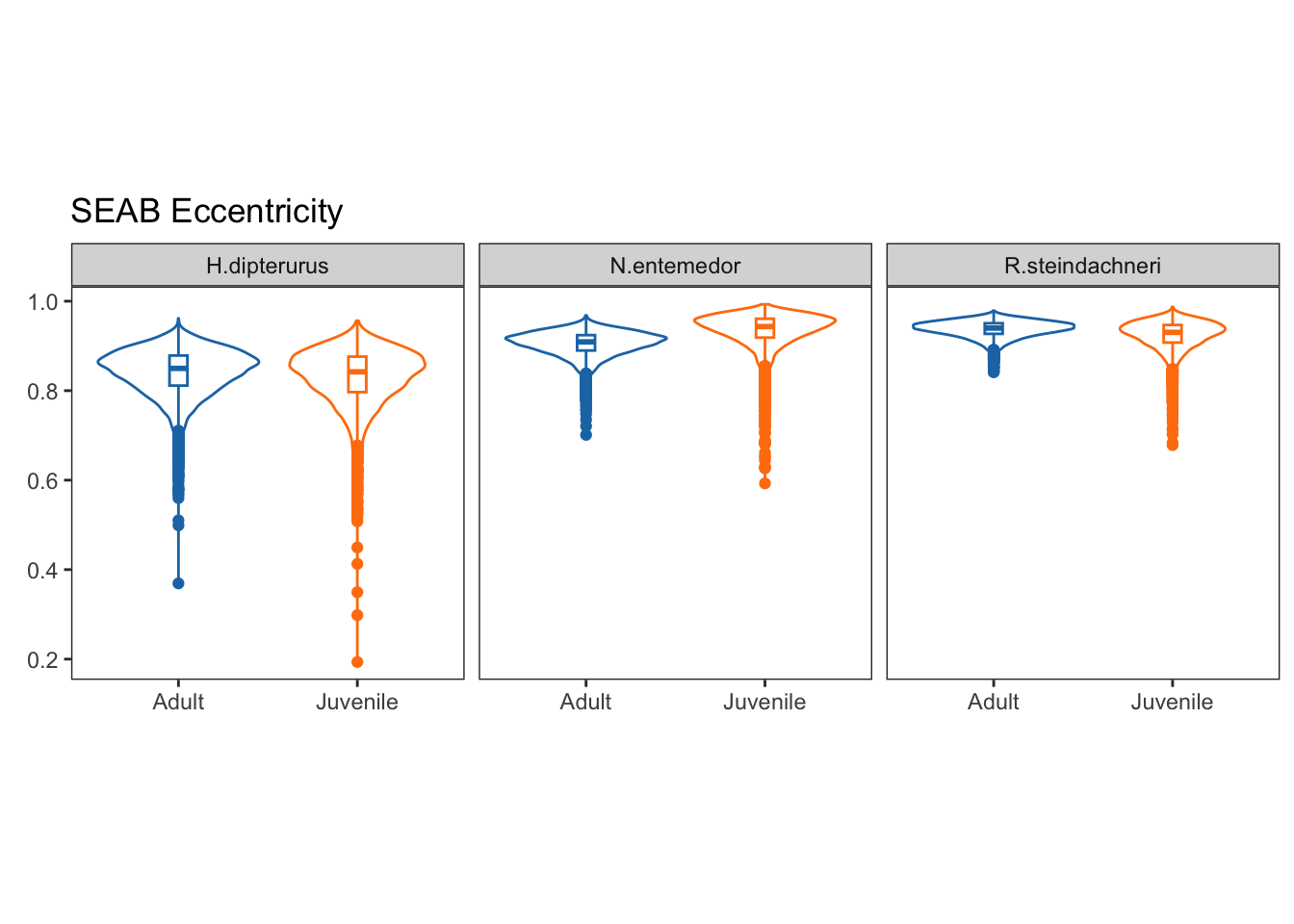
Theta:
th <- ecc %>% group_by(sp, j) %>% summarise(IQR.lower = hdi(theta, credMass = 0.5)[1],
IQR.upper = hdi(theta, credMass = 0.5)[2],
hdi.lower = hdi(theta)[1],
hdi.upper = hdi(theta)[2],
mean = mean(theta))`summarise()` has grouped output by 'sp'. You can override using the `.groups`
argument.thggplot(data = ecc, aes(x = j, y = theta, group = j, color = factor(j))) +
geom_violin(fill = NA, show.legend = F) +
#geom_point(aes(x = sp, y = mean), data = ag, show.legend = F) +
#geom_errorbar(aes(x = sp, ymin = IQ.lower, ymax = IQ.upper, group = sp), data = ag) +
geom_boxplot(width = 0.1, show.legend = F) +
theme_bw() +
labs(title = "SEAB theta",
x = element_blank(),
y = element_blank()) +
scale_color_manual(values = global_colors) +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.15, 0.1),
legend.background = element_blank()) +
facet_wrap(~sp)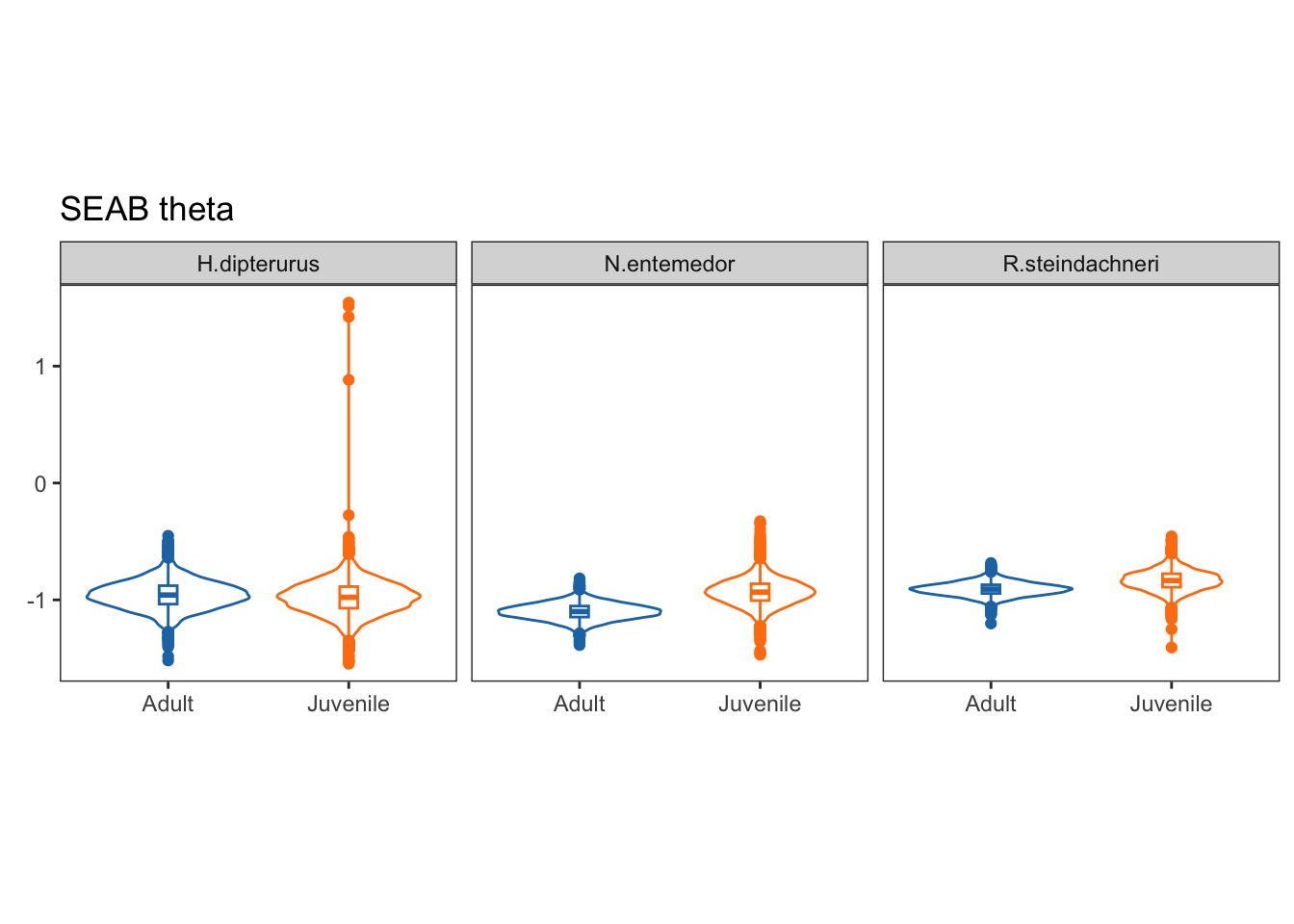
3.4.4 Season
lbls <- c("Warm", "Cold")
season_siber <- custom_siber(full_set, var_col = "Season", lbls, sp_col = "Species",
colors = global_colors, gp_label = "Season",
n.burnin = 750000, n.iter = 10000) Too few points to calculate an ellipseWarning: Removed 1 row containing missing values (`geom_path()`).Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 63
Unobserved stochastic nodes: 3
Total graph size: 78
Initializing model
Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 18
Unobserved stochastic nodes: 3
Total graph size: 33
Initializing modelNo id variables; using all as measure variablesCompiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 15
Unobserved stochastic nodes: 3
Total graph size: 30
Initializing model
Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 54
Unobserved stochastic nodes: 3
Total graph size: 69
Initializing modelNo id variables; using all as measure variablesWarning in createSiberObject(subdata[subdata[, sp_col] == species[i], 3:6]): At least one of your groups has less than 5 observations.
The absolute minimum sample size for each group is 3 in order
for the various ellipses and corresponding metrics to be
calculated. More reasonably though, a minimum of 5 data points
are required to calculate the two means and the 2x2 covariance
matrix and not run out of degrees of freedom. Check the item
named 'sample.sizes' in the object returned by this function
in order to locate the offending group. Bear in mind that NAs in
the sample.size matrix simply indicate groups that are not
present in that community, and is an acceptable data structure
for these analyses.Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 71
Unobserved stochastic nodes: 3
Total graph size: 86
Initializing model
Compiling model graph
Resolving undeclared variables
Allocating nodes
Graph information:
Observed stochastic nodes: 3
Unobserved stochastic nodes: 3
Total graph size: 18
Initializing modelNo id variables; using all as measure variables[1] "All models have been run. Check your working directory for the results"Plots of ellipses:
season_siber$ellips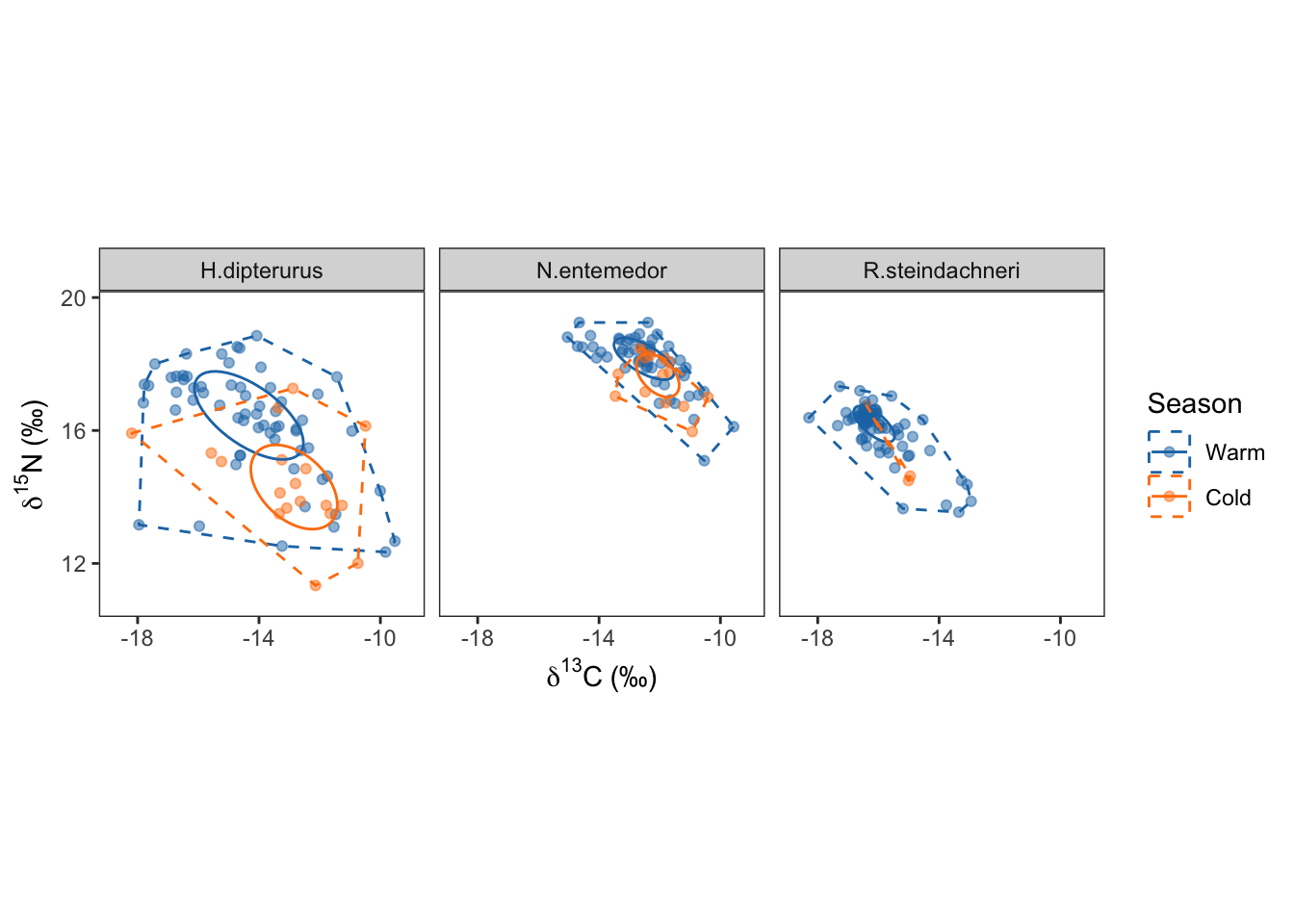
ecc <- data.frame()
for (sp in seq_along(species)) {
for (j in seq_along(lbls)) {
ecc <- rbind(ecc,
data.frame(sp = species[sp],
j = lbls[j],
ecc = apply(season_siber$ellips[[species[sp]]][[j]][,1:4], 1, mat_ecc),
theta = apply(season_siber$ellips[[species[sp]]][[j]][,1:4], 1, mat_theta)))
}
}ec <- ecc %>% group_by(sp, j) %>% summarise(IQR.lower = hdi(ecc, credMass = 0.5)[1],
IQR.upper = hdi(ecc, credMass = 0.5)[2],
hdi.lower = hdi(ecc)[1],
hdi.upper = hdi(ecc)[2],
mean = mean(ecc))`summarise()` has grouped output by 'sp'. You can override using the `.groups`
argument.ecggplot(data = ecc, aes(x = j, y = ecc, group = j, color = factor(j))) +
geom_violin(fill = NA, show.legend = F) +
#geom_point(aes(x = sp, y = mean), data = ag, show.legend = F) +
#geom_errorbar(aes(x = sp, ymin = IQ.lower, ymax = IQ.upper, group = sp), data = ag) +
geom_boxplot(width = 0.1, show.legend = F) +
theme_bw() +
labs(title = "SEAB Eccentricity",
x = element_blank(),
y = element_blank()) +
scale_color_manual(values = global_colors) +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.15, 0.1),
legend.background = element_blank()) +
facet_wrap(~sp)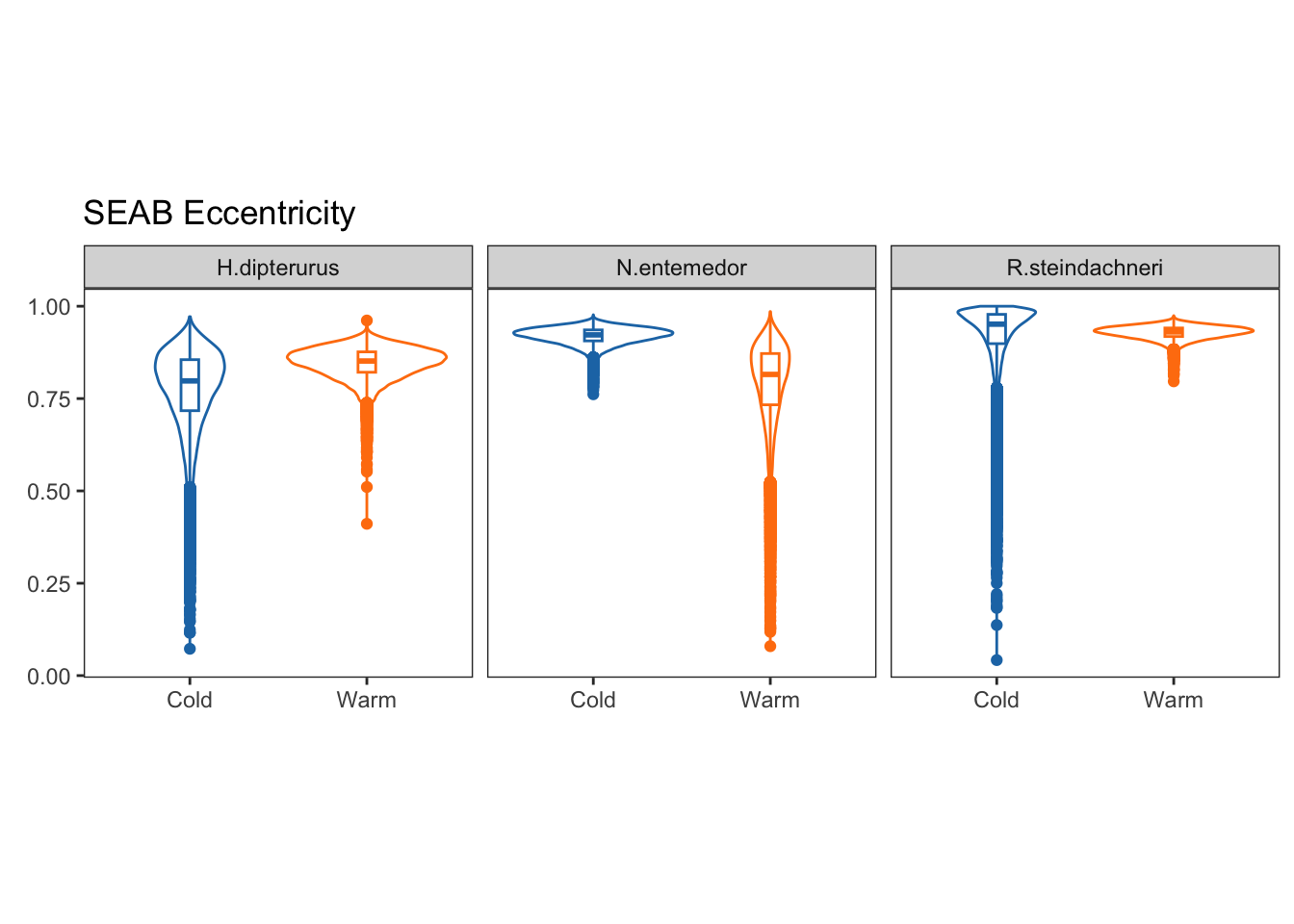
Theta:
th <- ecc %>% group_by(sp, j) %>% summarise(IQR.lower = hdi(theta, credMass = 0.5)[1],
IQR.upper = hdi(theta, credMass = 0.5)[2],
hdi.lower = hdi(theta)[1],
hdi.upper = hdi(theta)[2],
mean = mean(theta))`summarise()` has grouped output by 'sp'. You can override using the `.groups`
argument.thggplot(data = ecc, aes(x = j, y = theta, group = j, color = factor(j))) +
geom_violin(fill = NA, show.legend = F) +
#geom_point(aes(x = sp, y = mean), data = ag, show.legend = F) +
#geom_errorbar(aes(x = sp, ymin = IQ.lower, ymax = IQ.upper, group = sp), data = ag) +
geom_boxplot(width = 0.1, show.legend = F) +
theme_bw() +
labs(title = "SEAB theta",
x = element_blank(),
y = element_blank()) +
scale_color_manual(values = global_colors) +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.15, 0.1),
legend.background = element_blank()) +
facet_wrap(~sp)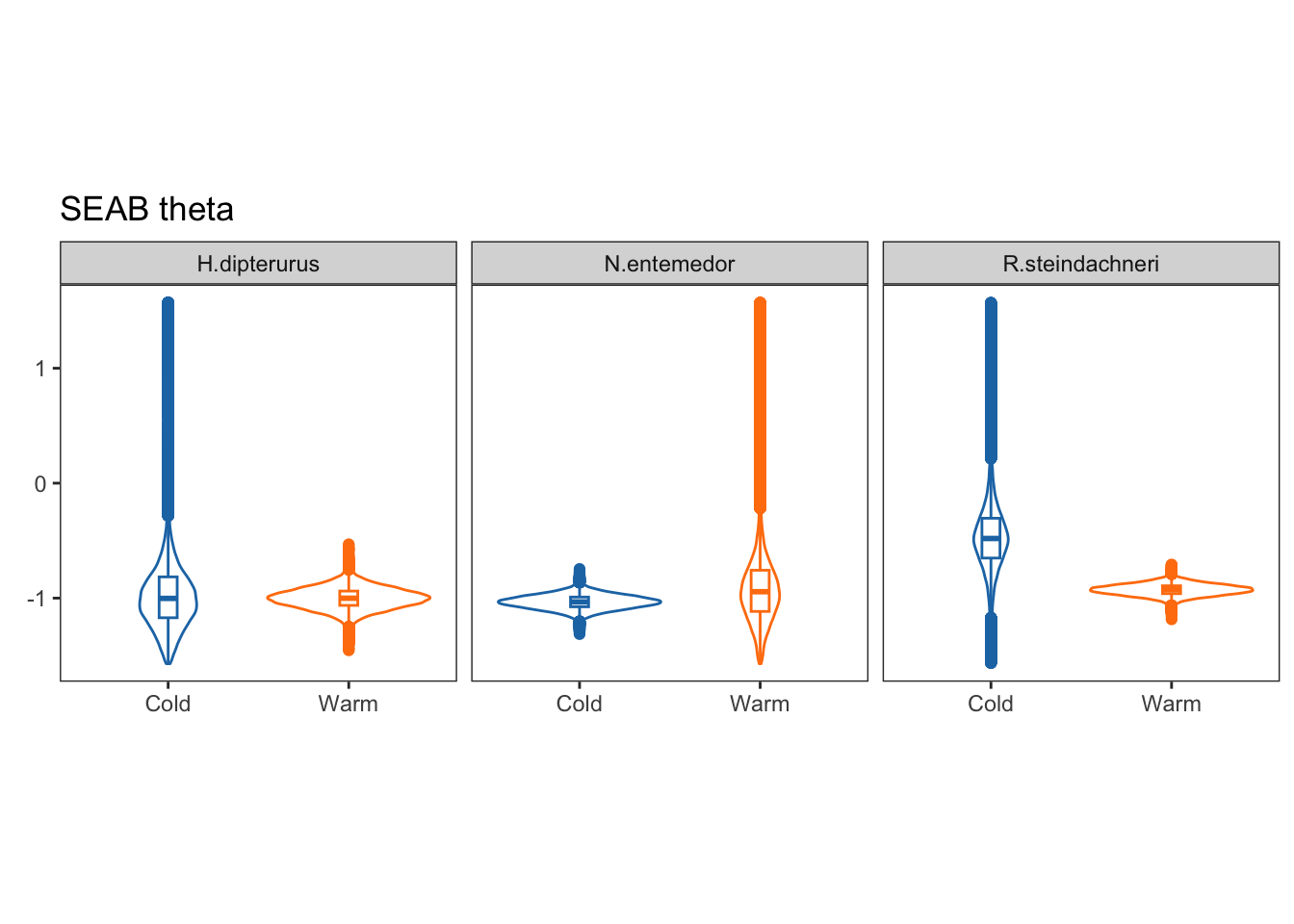
3.4.5 Final forestplot:
forest_data <- rbind(age_siber[[6]], season_siber[[6]], sex_siber[[6]])
forest <- ggplot(data = forest_data) +
geom_vline(xintercept = 0, linetype = "dashed", color = "#C9C8C8", size = 0.5) +
geom_linerange(aes(y = comp, xmin = IQr.lower, xmax = IQr.upper),
color = global_colors[1], size = 1) +
geom_linerange(aes(y = comp, xmin = hdi.lower, xmax = hdi.upper),
color = global_colors[1]) +
geom_point(aes(x = mean, y = comp),
color = global_colors[1], fill = "white", size = 1.5, shape = 21) +
theme_bw() +
theme(aspect.ratio = 1,
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(0.2, 0.15),
legend.background = element_blank()) +
scale_x_continuous(breaks = scales::pretty_breaks(n = 3)) +
labs(title = element_blank(),
x = element_blank(),
y = element_blank()) + facet_wrap(~sp)
# cairo_pdf("output/SIBER/forest_cats.pdf", family = "Times", width = 4, height = 2)
forest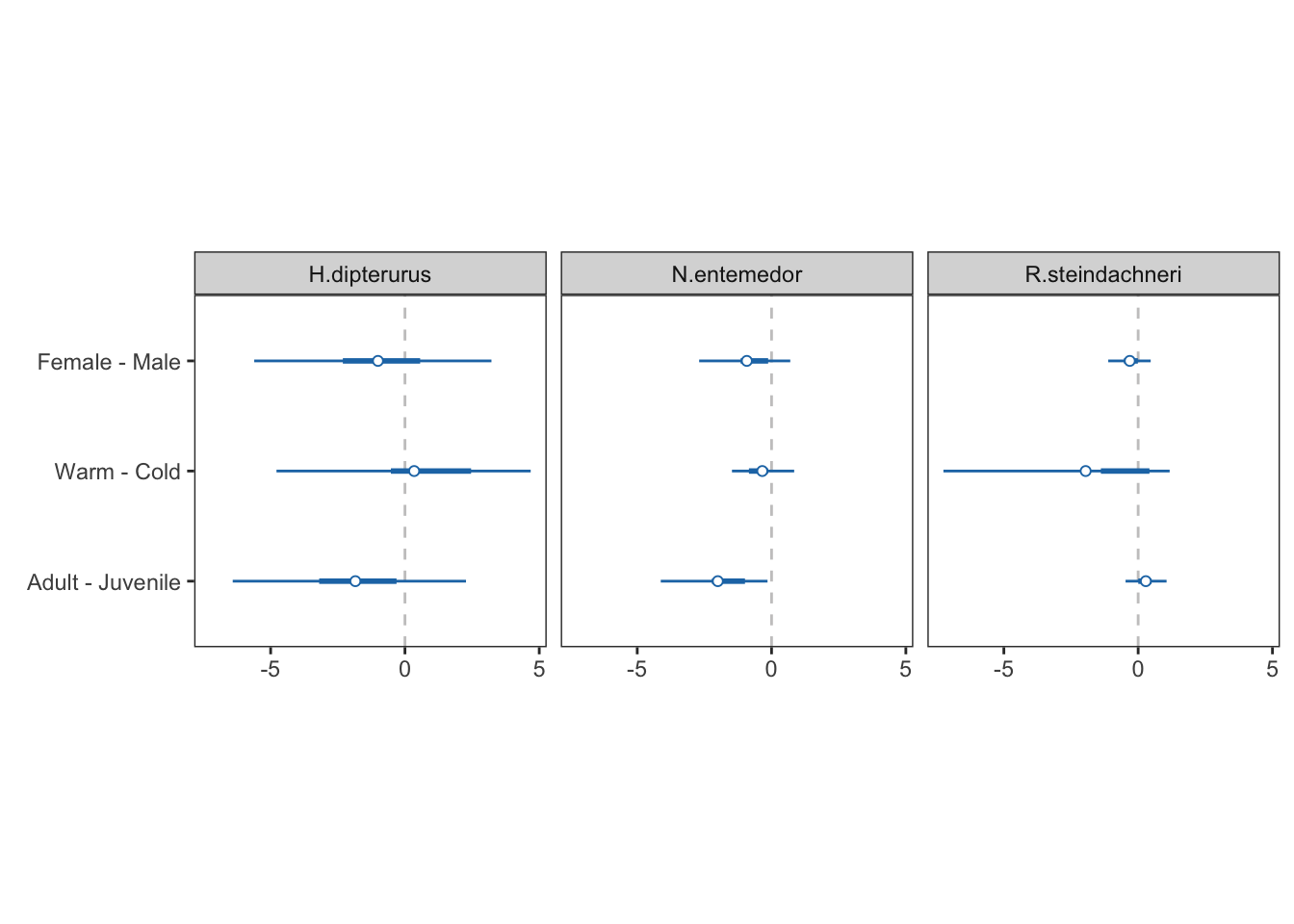
# dev.off()Tabular results:
Seasons:
season_siber$diff_glob[,c("comp", "sp", "mean")]season_siber$p_supSexes:
sex_siber$diff_glob[,c("comp", "sp", "mean")]sex_siber$p_supMaturity stages:
age_siber$diff_glob[,c("comp", "sp", "mean")]age_siber$p_sup